1. WHO. WHO | Ova le mamafa ma le puta. www.who.int/gho/ncd/
risk_factors/overweight/en/index.html. Accessed 29 January 2015.
2. Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery.
Am J Hum Genet. 2012;90:7�24.
3. Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic
studies of body mass index yield new insights for obesity biology. Nature.
2015; 518: 197--206.
4. Ling C, Del Guerra S, Lupi R, R�nn T, Granhall C, Luthman H, et al.
Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and
effect on insulin secretion. Diabetologia. 2008;51:615�22.
5. Van Dijk SJ, Molloy PL, Varinli H, Morrison JL, Muhlhausler BS. Epigenetics
and human obesity. Int J Obes (Lond). 2015;39:85�97.
6. Teh AL, Pan H, Chen L, Ong M-L, Dogra S, Wong J, et al. The effect of
genotype and in utero environment on interindividual variation in neonate
DNA methylomes. Genome Res. 2014;24:1064�74.
7. Olsson AH, Volkov P, Bacos K, Dayeh T, Hall E, Nilsson EA, et al. Genomewide
associations between genetic and epigenetic variation influence
mRNA expression and insulin secretion in human pancreatic islets. PLoS
Genet. 2014;10:e1004735.
8. Grundberg E, Meduri E, Sandling JK, Hedman AK, Keildson S, Buil A, et al.
Global analysis of DNA methylation variation in adipose tissue from twins
reveals links to disease-associated variants in distal regulatory elements.
Am J Hum Genet. 2013;93:876�90.
9. Ronn T, Volkov P, Gillberg L, Kokosar M, Perfilyev A, Jacobsen AL, et al.
Impact of age, BMI and HbA1c levels on the genome-wide DNA
methylation and mRNA expression patterns in human adipose tissue
and identification of epigenetic biomarkers in blood. Hum Mol Genet.
2015; 24: 3792--813.
10. Waterland RA, Michels KB. Epigenetic epidemiology of the developmental
origins hypothesis. Annu Rev Nutr. 2007;27:363�88.
11. McMillen IC, Rattanatray L, Duffield JA, Morrison JL, MacLaughlin SM, Gentili
S, et al. The early origins of later obesity: pathways and mechanisms. Adv
Exp Med Biol. 2009;646:71�81.
12. Ravelli A, van der Meulen J, Michels R, Osmond C, Barker D, Hales C, et al.
Glucose tolerance in adults after prenatal exposure to famine. Lancet.
1998; 351: 173--7.
13. McMillen IC, MacLaughlin SM, Muhlhausler BS, Gentili S, Duffield JL,
Morrison JL. Developmental origins of adult health and disease: the role of
periconceptional and foetal nutrition. Basic Clin Pharmacol Toxicol.
2008; 102: 82--9.
14. Zhang S, Rattanatray L, McMillen IC, Suter CM, Morrison JL. Periconceptional
nutrition and the early programming of a life of obesity or adversity. Prog
Biophys Mol Biol. 2011;106:307�14.
15. Bouret S, Levin BE, Ozanne SE. Gene-environment interactions controlling
energy and glucose homeostasis and the developmental origins of obesity.
Physiol Rev. 2015;95:47�82.
16. Borengasser SJ, Zhong Y, Kang P, Lindsey F, Ronis MJ, Badger TM, et al.
Maternal obesity enhances white adipose tissue differentiation and alters
genome-scale DNA methylation in male rat offspring. Endocrinology.
2013; 154: 4113--25.
17. Gluckman PD, Lillycrop KA, Vickers MH, Pleasants AB, Phillips ES, Beedle AS,
et al. Metabolic plasticity during mammalian development is directionally
dependent on early nutritional status. Proc Natl Acad Sci U S A.
2007; 104: 12796--800.
18. Godfrey KM, Sheppard A, Gluckman PD, Lillycrop KA, Burdge GC, McLean C,
et al. Epigenetic gene promoter methylation at birth is associated with
child�s later adiposity. Diabetes. 2011;60:1528�34.
19. McMillen IC, Adam CL, Muhlhausler BS. Early origins of obesity:
programming the appetite regulatory system. J Physiol. 2005;565(Pt 1):9�17.
20. Begum G, Stevens A, Smith EB, Connor K, Challis JR, Bloomfield F, et al.
Epigenetic changes in fetal hypothalamic energy regulating pathways are
associated with maternal undernutrition and twinning. FASEB J.
2012; 26: 1694--703.
21. Ge ZJ, Liang QX, Hou Y, Han ZM, Schatten H, Sun QY, et al. Maternal obesity
and diabetes may cause DNA methylation alteration in the spermatozoa of
offspring in mice. Reprod Biol Endocrinol. 2014;12:29.
22. Jousse C, Parry L, Lambert-Langlais S, Maurin AC, Averous J, Bruhat A, et al.
Perinatal undernutrition affects the methylation and expression of the leptin
gene in adults: implication for the understanding of metabolic syndrome.
FASEB J. 2011;25:3271�8.
23. Lan X, Cretney EC, Kropp J, Khateeb K, Berg MA, Penagaricano F, et al.
Maternal diet during pregnancy induces gene expression and DNA
methylation changes in fetal tissues in sheep. Front Genet. 2013;4:49.
24. Li CC, Young PE, Maloney CA, Eaton SA, Cowley MJ, Buckland ME, et al.
Maternal obesity and diabetes induces latent metabolic defects and
widespread epigenetic changes in isogenic mice. Epigenetics. 2013;8:602�11.
25. Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein
restriction of pregnant rats induces and folic acid supplementation prevents
epigenetic modification of hepatic gene expression in the offspring. J Nutr.
2005; 135: 1382--6.
26. Radford EJ, Ito M, Shi H, Corish JA, Yamazawa K, Isganaitis E, et al. In utero
effects. In utero undernourishment perturbs the adult sperm methylome
and intergenerational metabolism. Science. 2014;345(80):1255903.
27. Suter M, Bocock P, Showalter L, Hu M, Shope C, McKnight R, et al.
Epigenomics: maternal high-fat diet exposure in utero disrupts
peripheral circadian gene expression in nonhuman primates. FASEB J.
2011; 25: 714--26.
28. Suter MA, Ma J, Vuguin PM, Hartil K, Fiallo A, Harris RA, et al. In utero
exposure to a maternal high-fat diet alters the epigenetic histone code in a
murine model. Am J Obs Gynecol. 2014;210:463 e1�463 e11.
29. Tosh DN, Fu Q, Callaway CW, McKnight RA, McMillen IC, Ross MG, et al.
Epigenetics of programmed obesity: alteration in IUGR rat hepatic IGF1
mRNA expression and histone structure in rapid vs. delayed postnatal
catch-up growth. Am J Physiol Gastrointest Liver Physiol.
2010;299:G1023�9.
30. Sandovici I, Smith NH, Nitert MD, Ackers-Johnson M, Uribe-Lewis S, Ito Y,
et al. Maternal diet and aging alter the epigenetic control of a promoterenhancer
interaction at the Hnf4a gene in rat pancreatic islets. Proc Natl
Acad Sci U S A. 2011;108:5449�54.
31. Braunschweig M, Jagannathan V, Gutzwiller A, Bee G. Investigations on
transgenerational epigenetic response down the male line in F2 pigs. PLoS
One. 2012;7, e30583.
32. Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, et al. Paternally
induced transgenerational environmental reprogramming of metabolic
gene expression in mammals. Cell. 2010;143:1084�96.
33. Ost A, Lempradl A, Casas E, Weigert M, Tiko T, Deniz M, et al. Paternal diet
defines offspring chromatin state and intergenerational obesity. Cell.
2014; 159: 1352--64.
34. Mart�nez D, Pentinat T, Rib� S, Daviaud C, Bloks VW, Cebri� J, et al. In utero
undernutrition in male mice programs liver lipid metabolism in the secondgeneration
offspring involving altered Lxra DNA methylation. Cell Metab.
2014; 19: 941--51.
35. Wei Y, Yang C-R, Wei Y-P, Zhao Z-A, Hou Y, Schatten H, et al. Paternally
induced transgenerational inheritance of susceptibility to diabetes in
mammals. Proc Natl Acad Sci U S A. 2014;111:1873�8.
36. Grossniklaus U, Kelly WG, Kelly B, Ferguson-Smith AC, Pembrey M, Lindquist
S. Transgenerational epigenetic inheritance: how important is it? Nat Rev
Genet. 2013;14:228�35.
37. Pembrey M, Saffery R, Bygren LO. Human transgenerational responses to
early-life experience: potential impact on development, health and
biomedical research. J Med Genet. 2014;51:563�72.
38. Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl
supplements affect agouti gene expression in Avy/a mice. FASEB J.
1998; 12: 949--57.
39. Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility.
Nat Rev Genet. 2007;8:253�62.
40. Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at
the agouti locus in the mouse. Nat Genet. 1999;23:314�8.
41. Cropley JE, Suter CM, Beckman KB, Martin DI. Germ-line epigenetic
modification of the murine A vy allele by nutritional supplementation. Proc
Natl Acad Sci U S A. 2006;103:17308�12.
42. Hoile SP, Lillycrop KA, Thomas NA, Hanson MA, Burdge GC. Dietary protein
restriction during F0 pregnancy in rats induces transgenerational changes in
the hepatic transcriptome in female offspring. PLoS One. 2011;6, e21668.
43. Multhaup ML, Seldin MM, Jaffe AE, Lei X, Kirchner H, Mondal P, et al. Mousehuman
experimental epigenetic analysis unmasks dietary targets and
genetic liability for diabetic phenotypes. Cell Metab. 2015;21:138�49.
44. Michels KB, Binder AM, Dedeurwaerder S, Epstein CB, Greally JM, Gut I, et al.
Recommendations for the design and analysis of epigenome-wide
association studies. Nat Methods. 2013;10:949�55.
45. Dayeh TA, Olsson AH, Volkov P, Almgren P, R�nn T, Ling C. Identification of
CpG-SNPs associated with type 2 diabetes and differential DNA methylation
in human pancreatic islets. Diabetologia. 2013;56:1036�46.
46. Relton CL, Davey Smith G. Two-step epigenetic Mendelian randomization: a
strategy for establishing the causal role of epigenetic processes in pathways
to disease. Int J Epidemiol. 2012;41:161�76.
47. Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, et al.
Epigenome-wide association data implicate DNA methylation as an
intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol.
2013; 31: 142--7.
48. Yuan W, Xia Y, Bell CG, Yet I, Ferreira T, Ward KJ, et al. An integrated
epigenomic analysis for type 2 diabetes susceptibility loci in monozygotic
twins. Nat Commun. 2014;5:5719.
49. Nitert MD, Dayeh T, Volkov P, Elgzyri T, Hall E, Nilsson E, et al. Impact of an
exercise intervention on DNA methylation in skeletal muscle from firstdegree
relatives of patients with type 2 diabetes. Diabetes. 2012;61:3322�32.
50. Gagnon F, A�ssi D, Carri� A, Morange P-E, Tr�gou�t D-A. Robust validation of
methylation levels association at CPT1A locus with lipid plasma levels.
J Lipid Res. 2014;55:1189�91.
51. Demerath EW, Guan W, Grove ML, Aslibekyan S, Mendelson M, Zhou Y-H,
et al. Epigenome-wide association atudy (EWAS) of BMI, BMI change, and
waist circumference in African American adults identifies multiple replicated
loci. Hum Mol Genet. 2015:ddv161�.
52. Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, A�ssi D, Wahl S, et al. DNA
methylation and body-mass index: a genome-wide analysis. Lancet.
2014; 6736: 1--9.
53. Su S, Zhu H, Xu X, Wang X, Dong Y, Kapuku G, et al. DNA methylation of
the LY86 gene is associated with obesity, insulin resistance, and
inflammation. Twin Res Hum Genet. 2014;17:183�91.
54. Clarke-Harris R, Wilkin TJ, Hosking J, Pinkney J, Jeffery AN, Metcalf BS, et al.
PGC1? promoter methylation in blood at 5�7 years predicts adiposity from
9 to 14 years (EarlyBird 50). Diabetes. 2014;63:2528�37.
55. Guay S-P, Brisson D, Lamarche B, Biron S, Lescelleur O, Biertho L, et al.
ADRB3 gene promoter DNA methylation in blood and visceral adipose
tissue is associated with metabolic disturbances in men. Epigenomics.
2014; 6: 33--43.
56. Agha G, Houseman EA, Kelsey KT, Eaton CB, Buka SL, Loucks EB. Adiposity is
associated with DNA methylation profile in adipose tissue. Int J Epidemiol.
2014: 1--11.
57. Irvin MR, Zhi D, Joehanes R, Mendelson M, Aslibekyan S, Claas SA, et al.
Epigenome-wide association study of fasting blood lipids in the genetics of
lipid-lowering drugs and diet network study. Circulation. 2014;130:565�72.
58. Frazier-Wood AC, Aslibekyan S, Absher DM, Hopkins PN, Sha J, Tsai MY, et al.
Methylation at CPT1A locus is associated with lipoprotein subfraction
profiles. J Lipid Res. 2014;55:1324�30.
59. Pfeifferm L, Wahl S, Pilling LC, Reischl E, Sandling JK, Kunze S, et al. DNA
methylation of lipid-related genes affects blood lipid levels. Circ Cardiovasc
Genet. 2015.
60. Petersen A-K, Zeilinger S, Kastenm�ller G, R�misch-Margl W, Brugger M, Peters
A, et al. Epigenetics meets metabolomics: an epigenome-wide association
study with blood serum metabolic traits. Hum Mol Genet. 2014;23:534�45.
61. Hidalgo B, Irvin MR, Sha J, Zhi D, Aslibekyan S, Absher D, et al. Epigenomewide
association study of fasting measures of glucose, insulin, and HOMA-IR
in the genetics of lipid lowering drugs and diet network study. Diabetes.
2014; 63: 801--7.
62. Dayeh T, Volkov P, Sal� S, Hall E, Nilsson E, Olsson AH, et al. Genome-wide
DNA methylation analysis of human pancreatic islets from type 2 diabetic
and non-diabetic donors identifies candidate genes that influence insulin
secretion. PLoS Genet. 2014;10, e1004160.
63. Nilsson E, Jansson PA, Perfilyev A, Volkov P, Pedersen M, Svensson MK, et al.
Altered DNA methylation and differential expression of genes influencing
metabolism and inflammation in adipose tissue from subjects with type 2
diabetes. Diabetes. 2014;63:2962�76.
64. Benton MC, Johnstone A, Eccles D, Harmon B, Hayes MT, Lea RA, et al. An analysis of DNA methylation in human adipose tissue reveals differential modification of obesity genes before and after gastric bypass and weight
loss. Gene. 2015;16:1�21.
65. Bateson P, Gluckman P. Plasticity and robustness in development and
evolution. Int J Epidemiol. 2012;41:219�23.
66. Feinberg AP, Irizarry RA, Feinberg AP, Irizarry RA. Evolution in health and
medicine Sackler colloquium: stochastic epigenetic variation as a driving
force of development, evolutionary adaptation, and disease. Proc Natl Acad
Sci U S A. 2010;107(Suppl):1757�64.
67. Martino D, Loke YJ, Gordon L, Ollikainen M, Cruickshank MN, Saffery R, et al.
Longitudinal, genome-scale analysis of DNA methylation in twins from birth
to 18 months of age reveals rapid epigenetic change in early life and pairspecific
effects of discordance. Genome Biol. 2013;14:R42.
68. Tobi EW, Goeman JJ, Monajemi R, Gu H, Putter H, Zhang Y, et al. DNA
methylation signatures link prenatal famine exposure to growth and
metabolism. Nat Commun. 2014;5:5592.
69. Dominguez-Salas P, Moore SE, Baker MS, Bergen AW, Cox SE, Dyer RA, et al.
Maternal nutrition at conception modulates DNA methylation of human
metastable epialleles. Nat Commun. 2014;5:3746.
70. Quilter CR, Cooper WN, Cliffe KM, Skinner BM, Prentice PM, Nelson L, et al.
Impact on offspring methylation patterns of maternal gestational diabetes
mellitus and intrauterine growth restraint suggest common genes and
pathways linked to subsequent type 2 diabetes risk. FASEB J. 2014:1�12.
71. Morales E, Groom A, Lawlor DA, Relton CL. DNA methylation signatures in
cord blood associated with maternal gestational weight gain: results from
the ALSPAC cohort. BMC Res Notes. 2014;7:278.
72. Ruchat SM, Houde AA, Voisin G, St-Pierre J, Perron P, Baillargeon JP, et al.
Gestational diabetes mellitus epigenetically affects genes predominantly
involved in metabolic diseases. Epigenetics. 2013;8:935�43.
73. Liu X, Chen Q, Tsai H-J, Wang G, Hong X, Zhou Y, et al. Maternal
preconception body mass index and offspring cord blood DNA
methylation: exploration of early life origins of disease. Environ Mol
Mutagen. 2014;55:223�30.
74. Soubry A, Murphy SK, Wang F, Huang Z, Vidal AC, Fuemmeler BF, et al.
Newborns of obese parents have altered DNA methylation patterns at
imprinted genes. Int J Obes (Lond). 2015;39:650�7.
75. Jacobsen SC, Br�ns C, Bork-Jensen J, Ribel-Madsen R, Yang B, Lara E, et al.
Effects of short-term high-fat overfeeding on genome-wide DNA
methylation in the skeletal muscle of healthy young men. Diabetologia.
2012; 55: 3341--9.
76. Gillberg L, Jacobsen SC, R�nn T, Br�ns C, Vaag A. PPARGC1A DNA
methylation in subcutaneous adipose tissue in low birth weight subjects�
impact of 5 days of high-fat overfeeding. Metabolism. 2014;63:263�71.
77. Huang Y-T, Maccani JZJ, Hawley NL, Wing RR, Kelsey KT, McCaffery JM.
Epigenetic patterns in successful weight loss maintainers: a pilot study. Int J
Obes (Lond). 2015;39:865�8.
78. Barres R, Kirchner H, Rasmussen M, Yan J, Kantor FR, Krook A, N�slund E,
Zierath JR. Weight loss after gastric bypass surgery in human obesity
remodels promoter methylation. Cell Rep. 2013:1�8.
79. Ahrens M, Ammerpohl O, von Sch�nfels W, Kolarova J, Bens S, Itzel T, et al.
DNA methylation analysis in nonalcoholic fatty liver disease suggests
distinct disease-specific and remodeling signatures after bariatric surgery.
Cell Metab. 2013;18:296�302.
80. Voisin S, Eynon N, Yan X, Bishop DJ. Exercise training and DNA methylation
in humans. Acta Physiol (Oxf). 2014;213:39�59.
81. Lindholm ME, Marabita F, Gomez-Cabrero D, Rundqvist H, Ekstr�m TJ,
Tegn�r J, et al. An integrative analysis reveals coordinated reprogramming
of the epigenome and the transcriptome in human skeletal muscle after
training. Epigenetics. 2014;9:1557�69.
82. Denham J, O�Brien BJ, Marques FZ, Charchar FJ. Changes in the leukocyte
methylome and its effect on cardiovascular related genes after exercise.
J Appl Physiol. 2014:jap.00878.2014.
83. Rowlands DS, Page RA, Sukala WR, Giri M, Ghimbovschi SD, Hayat I, et al.
Multi-omic integrated networks connect DNA methylation and miRNA with
skeletal muscle plasticity to chronic exercise in type 2 diabetic obesity.
Physiol Genomics. 2014;46:747�65.
84. Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schonfels W, Ahrens M,
et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad
Sci. 2014;111:15538�43.
85. Alm�n MS, Nilsson EK, Jacobsson JA, Kalnina I, Klovins J, Fredriksson R, et al.
Genome-wide analysis reveals DNA methylation markers that vary with
both age and obesity. Gene. 2014.;548:61�7
86. Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments
in analysis of DNA methylation data. Bioinformatics. 2014;30:1431�9.
87. Wells JC. A critical appraisal of the predictive adaptive response hypothesis.
Int J Epidemiol. 2012;41:229�35.
88. Williams-Wyss O, Zhang S, MacLaughlin SM, Kleemann D, Walker SK, Suter
CM, et al. Embryo number and periconceptional undernutrition in the
sheep have differential effects on adrenal epigenotype, growth, and
development. Am J Physiol Endocrinol Metab. 2014;307:E141�50.
89. Zhang S, Rattanatray L, Morrison JL, Nicholas LM, Lie S, McMillen IC.
Maternal obesity and the early origins of childhood obesity: weighing up
the benefits and costs of maternal weight loss in the periconceptional
period for the offspring. Exp Diabetes Res. 2011;2011:585749.
90. Zhang S, Williams-Wyss O, MacLaughlin SM, Walker SK, Kleemann DO, Suter
CM, et al. Maternal undernutrition during the first week after conception
results in decreased expression of glucocorticoid receptor mRNA in the
absence of GR exon 17 hypermethylation in the fetal pituitary in late
gestation. J Dev Orig Heal Dis. 2013;4:391�401.
91. Lie S, Morrison JL, Williams-Wyss O, Suter CM, Humphreys DT, Ozanne SE,
et al. Periconceptional undernutrition programs changes in insulin-signaling
molecules and microRNAs in skeletal muscle in singleton and twin fetal
sheep. Biol Reprod. 2014;90:5.
92. Van Straten EM, van Meer H, Huijkman NC, van Dijk TH, Baller JF, Verkade
HJ, et al. Fetal liver X receptor activation acutely induces lipogenesis but
does not affect plasma lipid response to a high-fat diet in adult mice. Am J
Physiol Endocrinol Metab. 2009;297:E1171�8.
93. Fernandez-Twinn DS, Alfaradhi MZ, Martin-Gronert MS, Duque-Guimaraes
DE, Piekarz A, Ferland-McCollough D, et al. Downregulation of IRS-1 in
adipose tissue of offspring of obese mice is programmed cellautonomously
through post-transcriptional mechanisms. Mol Metab.
2014; 3: 325--33.
94. Waterland RA, Travisano M, Tahiliani KG. Diet-induced hypermethylation at
agouti viable yellow is not inherited transgenerationally through the female.
FASEB J. 2007;21:3380�5.
95. Ge ZJ, Luo SM, Lin F, Liang QX, Huang L, Wei YC, et al. DNA methylation in
oocytes and liver of female mice and their offspring: effects of high-fat-dietinduced
obesity. Env Heal Perspect. 2014;122:159�64.
96. Ollikainen M, Ismail K, Gervin K, Kyll�nen A, Hakkarainen A, Lundbom J, et al.
Genome-wide blood DNA methylation alterations at regulatory elements
and heterochromatic regions in monozygotic twins discordant for obesity
and liver fat. Clin Epigenetics. 2015;7:1�13.








 O le tele o mea taua, faʻamaʻi tele, ma malamalama atili i auala e faʻavae ai fegalegaleaiga i le va o le soifuaga, siosiomaga, ma genetics e taua tele mo le atinaʻeina o fuafuaga lelei mo le puipuiga ma togafitiga [1].
O le tele o mea taua, faʻamaʻi tele, ma malamalama atili i auala e faʻavae ai fegalegaleaiga i le va o le soifuaga, siosiomaga, ma genetics e taua tele mo le atinaʻeina o fuafuaga lelei mo le puipuiga ma togafitiga [1]. Animal models provide unique opportunities for highly controlled studies that provide mechanistic insight into�the role of specific epigenetic marks, both as indicators of current metabolic status and as predictors of the future risk of obesity and metabolic disease. A particularly important aspect of animal studies is that they allow for the assessment of epigenetic changes within target tissues, including the liver and hypothalamus, which is much more difficult in humans. Moreover, the ability to harvest large quantities of fresh tissue makes it possible to assess multiple chromatin marks as well as DNA methylation. Some of these epigenetic modifications either alone or in combination may be responsive to environmental programming. In animal models, it is also possible to study multiple generations of offspring and thus enable differentiation between trans-generational and intergenerational transmission of obesity risk mediated by epigenetic memory of parental nutritional status, which cannot be easily distinguished in human studies. We use the former term for meiotic transmission of risk in the absence of continued exposure while the latter primarily entails direct transmission of risk through metabolic reprogramming of the fetus or gametes.
Animal models provide unique opportunities for highly controlled studies that provide mechanistic insight into�the role of specific epigenetic marks, both as indicators of current metabolic status and as predictors of the future risk of obesity and metabolic disease. A particularly important aspect of animal studies is that they allow for the assessment of epigenetic changes within target tissues, including the liver and hypothalamus, which is much more difficult in humans. Moreover, the ability to harvest large quantities of fresh tissue makes it possible to assess multiple chromatin marks as well as DNA methylation. Some of these epigenetic modifications either alone or in combination may be responsive to environmental programming. In animal models, it is also possible to study multiple generations of offspring and thus enable differentiation between trans-generational and intergenerational transmission of obesity risk mediated by epigenetic memory of parental nutritional status, which cannot be easily distinguished in human studies. We use the former term for meiotic transmission of risk in the absence of continued exposure while the latter primarily entails direct transmission of risk through metabolic reprogramming of the fetus or gametes.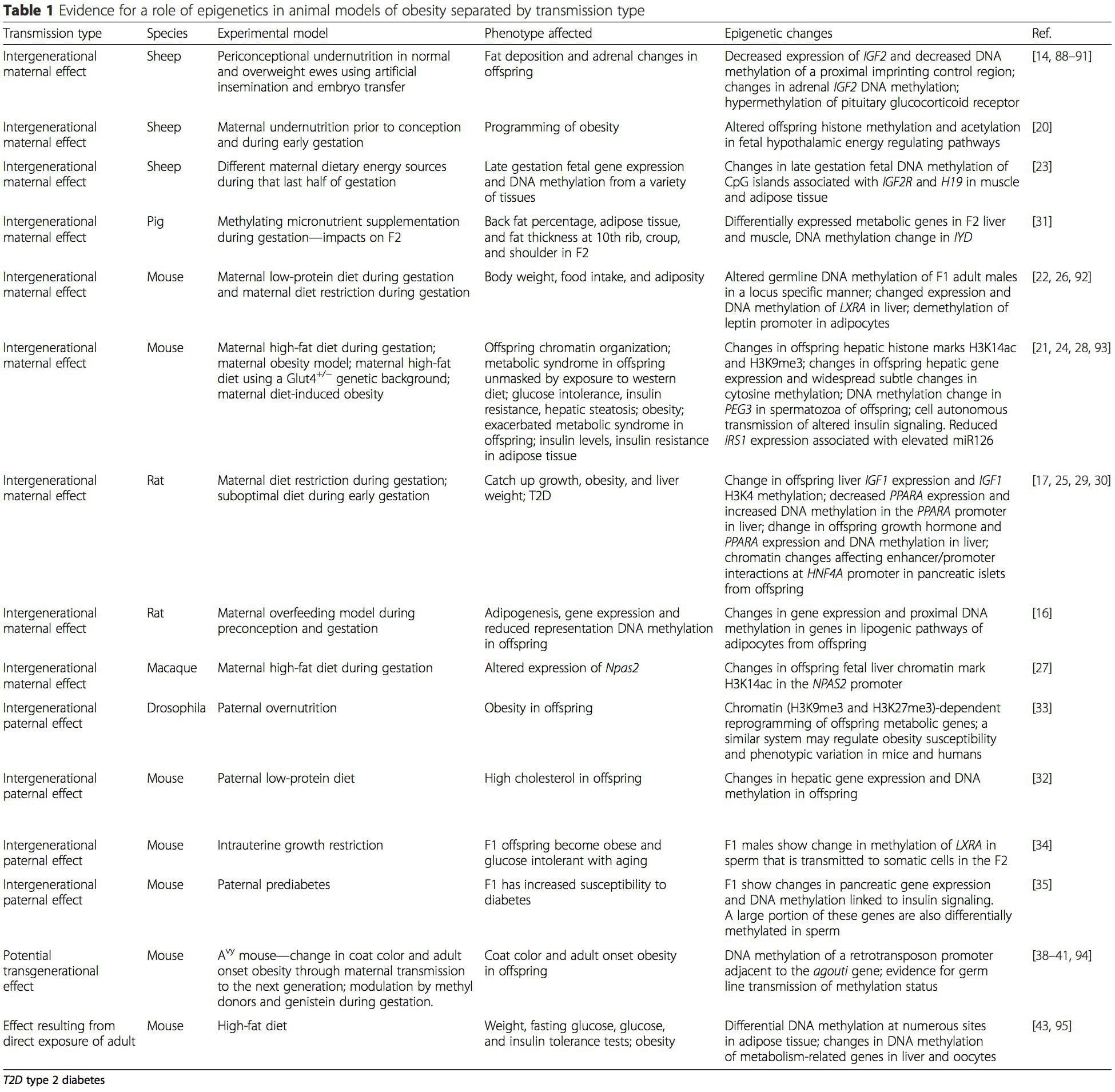 (i) Suiga o le Epigenetic i le Atinaʻe Faʻatasi ma Meaʻai Fafaga I le taimi o le Gestation
(i) Suiga o le Epigenetic i le Atinaʻe Faʻatasi ma Meaʻai Fafaga I le taimi o le Gestation Maternal nutritional supplementation, undernutrition, and over nutrition during pregnancy can alter fat deposition and energy homeostasis in offspring [11, 13�15, 19]. Associated with these effects in the offspring are changes in DNA methylation, histone post-translational modifications, and gene expression for several target genes,�especially genes regulating fatty acid metabolism and insulin signaling [16, 17, 20�30]. The diversity of animal models used in these studies and the common metabolic pathways impacted suggest an evolutionarily conserved adaptive response mediated by epigenetic modification. However, few of the specific identified genes and epigenetic changes have been cross-validated in related studies, and large-scale genome-wide investigations have typically not been applied. A major hindrance to comparison of these studies is the different develop mental windows subjected to nutritional challenge, which may cause considerably different outcomes. Proof that the epigenetic changes are causal rather than being associated with offspring phenotypic changes is also required. This will necessitate the identification of a parental nutritionally induced epigenetic �memory� response that precedes development of the altered phenotype in offspring.
Maternal nutritional supplementation, undernutrition, and over nutrition during pregnancy can alter fat deposition and energy homeostasis in offspring [11, 13�15, 19]. Associated with these effects in the offspring are changes in DNA methylation, histone post-translational modifications, and gene expression for several target genes,�especially genes regulating fatty acid metabolism and insulin signaling [16, 17, 20�30]. The diversity of animal models used in these studies and the common metabolic pathways impacted suggest an evolutionarily conserved adaptive response mediated by epigenetic modification. However, few of the specific identified genes and epigenetic changes have been cross-validated in related studies, and large-scale genome-wide investigations have typically not been applied. A major hindrance to comparison of these studies is the different develop mental windows subjected to nutritional challenge, which may cause considerably different outcomes. Proof that the epigenetic changes are causal rather than being associated with offspring phenotypic changes is also required. This will necessitate the identification of a parental nutritionally induced epigenetic �memory� response that precedes development of the altered phenotype in offspring. Emerging studies have demonstrated that paternal plane of nutrition can impact offspring fat deposition and epigenetic marks [31�34]. One recent investigation using mice has demonstrated that paternal pre-diabetes leads to increased susceptibility to diabetes in F1 offspring with associated changes in pancreatic gene expression and DNA methylation linked to insulin signaling [35]. Importantly, there was an overlap of these epigenetic changes in pancreatic islets and sperm suggesting germ line inheritance. However, most of these studies, although intriguing in their implications, are limited in the genomic scale of investigation and frequently show weak and somewhat transient epigenetic alterations associated with mild metabolic phenotypes in offspring.
Emerging studies have demonstrated that paternal plane of nutrition can impact offspring fat deposition and epigenetic marks [31�34]. One recent investigation using mice has demonstrated that paternal pre-diabetes leads to increased susceptibility to diabetes in F1 offspring with associated changes in pancreatic gene expression and DNA methylation linked to insulin signaling [35]. Importantly, there was an overlap of these epigenetic changes in pancreatic islets and sperm suggesting germ line inheritance. However, most of these studies, although intriguing in their implications, are limited in the genomic scale of investigation and frequently show weak and somewhat transient epigenetic alterations associated with mild metabolic phenotypes in offspring.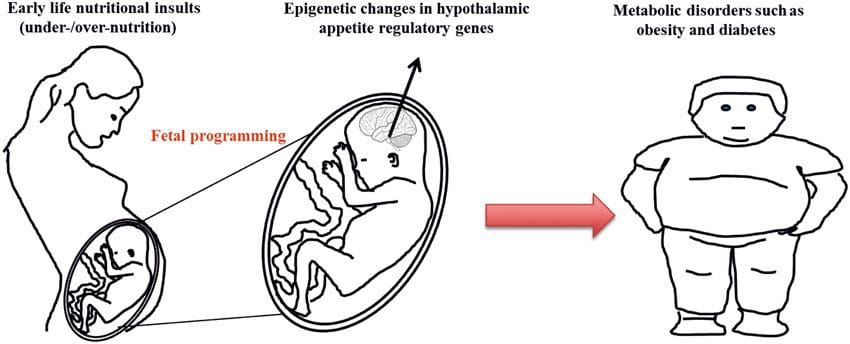 O le faʻaaogaina o faʻamaumauga o le epigenetic i le tele o augatupulaga e lelei ona faʻamatalaina i faʻalaʻau laau ma C. elegans, but its significance in mammals is still much debated [36, 37]. An epigenetic basis for grand- parental transmission of phenotypes in response to dietary exposures has been well established, including in livestock species [31]. The most influential studies demonstrating effects of epigenetic transmission impacting offspring phenotype have used the example of the viable yellow agouti (Avy) mouse [38]. In this mouse, an insertion of a retrotransposon upstream of the agouti gene causes its constitutive expression and consequent yellow coat color and adult onset obesity. Maternal transmission through the germ line results in DNA methylation�mediated silencing of agouti expression resulting in wild-type coat color and lean phenotype of the offspring [39, 40]. Importantly, subsequent studies in these mice demonstrated that maternal exposure to methyl donors causes a shift in coat color [41]. One study has reported transmission of a phenotype to the F3 generation and alterations in expression of large number of genes in response to protein restriction in F0 [42]; however, alterations in expression were highly variable and a direct link to epigenetic changes was not identified in this system.
O le faʻaaogaina o faʻamaumauga o le epigenetic i le tele o augatupulaga e lelei ona faʻamatalaina i faʻalaʻau laau ma C. elegans, but its significance in mammals is still much debated [36, 37]. An epigenetic basis for grand- parental transmission of phenotypes in response to dietary exposures has been well established, including in livestock species [31]. The most influential studies demonstrating effects of epigenetic transmission impacting offspring phenotype have used the example of the viable yellow agouti (Avy) mouse [38]. In this mouse, an insertion of a retrotransposon upstream of the agouti gene causes its constitutive expression and consequent yellow coat color and adult onset obesity. Maternal transmission through the germ line results in DNA methylation�mediated silencing of agouti expression resulting in wild-type coat color and lean phenotype of the offspring [39, 40]. Importantly, subsequent studies in these mice demonstrated that maternal exposure to methyl donors causes a shift in coat color [41]. One study has reported transmission of a phenotype to the F3 generation and alterations in expression of large number of genes in response to protein restriction in F0 [42]; however, alterations in expression were highly variable and a direct link to epigenetic changes was not identified in this system. E ui o le tele o suʻesuʻega ua faʻamalamalamaina suiga o le epigenetone i meaʻai, e faʻaaoga ai le faʻaaogaina o nofoaga faʻataʻatia a le au faipisinisi, o loʻo i ai ni nai suʻesuega lautele na faia. O se suʻesuʻega talu ai nei sa taulaʻi i le fuafuaina o le epigenethera faʻasolosolo o mea taumafa maualuga / taumafa-meaʻai i totonu o tamaʻi matutua e faʻaaoga ai genome-wide generic expression ma DNA methylation analyzes [43]. O lenei suʻesuʻega na faʻaalia ai 232 faʻapitoa methylated eria (DMRs) i adipocytes mai le pulea ma le maualuga-gaʻo fafagaina o nifo. O le mea sili ona taua, o vaega tutusa o tagata mo le DMR misa sa ese foi le methylated i le tino o le tino mai le faitau aofaʻi o tagata maualuluga ma vaivai, ma ua faʻamaonia ai le maoae o le faʻasaoina o nei vaipanoa. O lenei taunuʻuga e faʻamalosia ai le taua o le DMR ua faʻamaonia i le faʻatonutonuina o le malosiaga o le homeostasis i mamame.
E ui o le tele o suʻesuʻega ua faʻamalamalamaina suiga o le epigenetone i meaʻai, e faʻaaoga ai le faʻaaogaina o nofoaga faʻataʻatia a le au faipisinisi, o loʻo i ai ni nai suʻesuega lautele na faia. O se suʻesuʻega talu ai nei sa taulaʻi i le fuafuaina o le epigenethera faʻasolosolo o mea taumafa maualuga / taumafa-meaʻai i totonu o tamaʻi matutua e faʻaaoga ai genome-wide generic expression ma DNA methylation analyzes [43]. O lenei suʻesuʻega na faʻaalia ai 232 faʻapitoa methylated eria (DMRs) i adipocytes mai le pulea ma le maualuga-gaʻo fafagaina o nifo. O le mea sili ona taua, o vaega tutusa o tagata mo le DMR misa sa ese foi le methylated i le tino o le tino mai le faitau aofaʻi o tagata maualuluga ma vaivai, ma ua faʻamaonia ai le maoae o le faʻasaoina o nei vaipanoa. O lenei taunuʻuga e faʻamalosia ai le taua o le DMR ua faʻamaonia i le faʻatonutonuina o le malosiaga o le homeostasis i mamame.
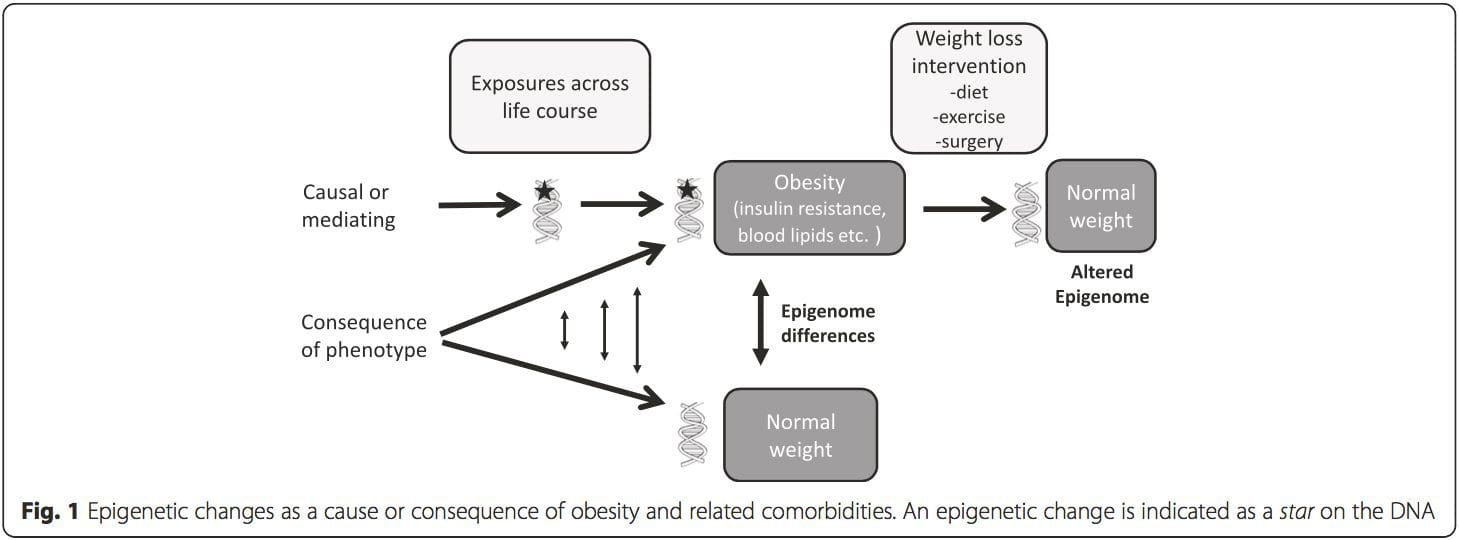 (i) Suʻesuʻega faʻatalanoaga faʻatasi. Genetic polymorphisms that are associated with an increased risk of developing particular conditions are a priori linked to the causative genes. The presence of differential�methylation in such regions infers functional relevance of these epigenetic changes in controlling expression of the proximal gene(s). There are strong cis-acting genetic effects underpinning much epigenetic variation [7, 45], and in population-based studies, methods that use genetic surrogates to infer a causal or mediating role of epigenome differences have been applied [7, 46�48]. The use of familial genetic information can also lead to the identification of potentially causative candidate regions showing phenotype-related differential methylation [49].
(i) Suʻesuʻega faʻatalanoaga faʻatasi. Genetic polymorphisms that are associated with an increased risk of developing particular conditions are a priori linked to the causative genes. The presence of differential�methylation in such regions infers functional relevance of these epigenetic changes in controlling expression of the proximal gene(s). There are strong cis-acting genetic effects underpinning much epigenetic variation [7, 45], and in population-based studies, methods that use genetic surrogates to infer a causal or mediating role of epigenome differences have been applied [7, 46�48]. The use of familial genetic information can also lead to the identification of potentially causative candidate regions showing phenotype-related differential methylation [49].
 Mai nei suʻesuʻega, o le methylation fesuiaiga o le PGC1A, HIF3A, ABCG1, ma le CPT1A ma le RXRA [18] na faʻamatalaina talu ai nei na faʻaalia mai o ni tagata e ola faʻatasi ma, pe atonu foi o le, o le soifua maloloina ole soifua maloloina e avea ma sui talafeagai mo le atiina ae o faamai pipisi. .
Mai nei suʻesuʻega, o le methylation fesuiaiga o le PGC1A, HIF3A, ABCG1, ma le CPT1A ma le RXRA [18] na faʻamatalaina talu ai nei na faʻaalia mai o ni tagata e ola faʻatasi ma, pe atonu foi o le, o le soifua maloloina ole soifua maloloina e avea ma sui talafeagai mo le atiina ae o faamai pipisi. . Epigenetic variation is highly influenced by the underlying genetic variation, with genotype estimated to explain ~20�40 % of the variation [6, 8]. Recently, a number of studies have begun to integrate methylome and genotype data to identify methylation quantitative trait loci (meQTL) associated with disease phenotypes. For instance, in adipose tissue, an meQTL overlapping�with a BMI genetic risk locus has been identified in an enhancer element upstream of ADCY3 [8]. Other studies have also identified overlaps between known obesity and T2DM risk loci and DMRs associated with obesity and T2DM [43, 48, 62]. Methylation of a number of such DMRs was also modulated by high-fat feeding in mice [43] and weight loss in humans [64]. These results identify an intriguing link between genetic variations linked with disease susceptibility and their association with regions of the genome that undergo epigenetic modifications in response to nutritional challenges, implying a causal relationship. The close connection between genetic and epigenetic variation may signify their essential roles in generating individual variation [65, 66]. However, while these findings suggest that DNA methylation may be a mediator of genetic effects, it is also important to consider that both genetic and epigenetic processes could act independently on the same genes. Twin studies [8, 63, 67] can provide important insights and indicate that inter-individual differences in levels of DNA methylation arise predominantly from non-shared environment and stochastic influences, minimally from shared environmental effects, but also with a significant impact of genetic variation.
Epigenetic variation is highly influenced by the underlying genetic variation, with genotype estimated to explain ~20�40 % of the variation [6, 8]. Recently, a number of studies have begun to integrate methylome and genotype data to identify methylation quantitative trait loci (meQTL) associated with disease phenotypes. For instance, in adipose tissue, an meQTL overlapping�with a BMI genetic risk locus has been identified in an enhancer element upstream of ADCY3 [8]. Other studies have also identified overlaps between known obesity and T2DM risk loci and DMRs associated with obesity and T2DM [43, 48, 62]. Methylation of a number of such DMRs was also modulated by high-fat feeding in mice [43] and weight loss in humans [64]. These results identify an intriguing link between genetic variations linked with disease susceptibility and their association with regions of the genome that undergo epigenetic modifications in response to nutritional challenges, implying a causal relationship. The close connection between genetic and epigenetic variation may signify their essential roles in generating individual variation [65, 66]. However, while these findings suggest that DNA methylation may be a mediator of genetic effects, it is also important to consider that both genetic and epigenetic processes could act independently on the same genes. Twin studies [8, 63, 67] can provide important insights and indicate that inter-individual differences in levels of DNA methylation arise predominantly from non-shared environment and stochastic influences, minimally from shared environmental effects, but also with a significant impact of genetic variation. Tausaga faʻapitoa: Two recently published studies made use of human populations that experienced �natural� variations in nutrient supply to study the impact of maternal nutrition before or during pregnancy on DNA methylation in the offspring [68, 69]. The first study used a Gambian mother-child cohort to show that both seasonal variations in maternal methyl donor intake during pregnancy and maternal pre-pregnancy BMI were associated with altered methylation in the infants [69]. The second study utilized adult offspring from the Dutch Hunger Winter cohort to investigate the effect of prenatal exposure to an acute period of severe maternal undernutrition on DNA methylation of genes involved in growth and metabolism in adulthood [68]. The results highlighted the importance of the timing of the exposure in its impact on the epigenome, since significant epigenetic effects were only identified in individuals exposed to famine during early gestation. Importantly, the epigenetic changes occurred in conjunction with increased BMI; however, it was not possible to establish in this study whether these changes were present earlier in life or a consequence of the higher BMI.
Tausaga faʻapitoa: Two recently published studies made use of human populations that experienced �natural� variations in nutrient supply to study the impact of maternal nutrition before or during pregnancy on DNA methylation in the offspring [68, 69]. The first study used a Gambian mother-child cohort to show that both seasonal variations in maternal methyl donor intake during pregnancy and maternal pre-pregnancy BMI were associated with altered methylation in the infants [69]. The second study utilized adult offspring from the Dutch Hunger Winter cohort to investigate the effect of prenatal exposure to an acute period of severe maternal undernutrition on DNA methylation of genes involved in growth and metabolism in adulthood [68]. The results highlighted the importance of the timing of the exposure in its impact on the epigenome, since significant epigenetic effects were only identified in individuals exposed to famine during early gestation. Importantly, the epigenetic changes occurred in conjunction with increased BMI; however, it was not possible to establish in this study whether these changes were present earlier in life or a consequence of the higher BMI. Siosiomaga Faʻasalalauga: The epigenome is established de novo during embryonic development, and therefore, the prenatal environment most likely has the most significant impact on the epigenome. However, it is now clear that changes do occur in the �mature� epigenome under the influence of a range of conditions, including aging, exposure to toxins, and dietary alterations. For example, changes in DNA methylation in numerous genes in skeletal muscle and PGC1A in adipose tissue have been demonstrated in response to a high-fat diet [75, 76]. Interventions to lose body fat mass have also been associated with changes in DNA methylation. Studies have reported that the DNA methylation profiles of adipose tissue [43, 64], peripheral blood mononuclear cells [77], and muscle tissue [78] in formerly obese patients become more similar to the profiles of lean subjects following weight loss. Weight loss surgery also partially reversed non-alcoholic fatty liver disease-associated methylation changes in liver [79] and in another study led to hypomethylation of multiple obesity candidate genes, with more pronounced effects in subcutaneous compared to omental (visceral) fat [64]. Accumulating evidence suggests that exercise interventions can also influence DNA methylation. Most of these studies have been conducted in lean individuals [80�82], but one exercise study in obese T2DM subjects also demonstrated changes in DNA methylation, including in genes involved in fatty acid and glucose transport [83]. Epigenetic changes also occur with aging, and recent data suggest a role of obesity in augmenting them [9, 84, 85]. Obesity accelerated the epigenetic age of liver tissue, but in contrast to the findings described above, this effect was not reversible after weight loss [84].
Siosiomaga Faʻasalalauga: The epigenome is established de novo during embryonic development, and therefore, the prenatal environment most likely has the most significant impact on the epigenome. However, it is now clear that changes do occur in the �mature� epigenome under the influence of a range of conditions, including aging, exposure to toxins, and dietary alterations. For example, changes in DNA methylation in numerous genes in skeletal muscle and PGC1A in adipose tissue have been demonstrated in response to a high-fat diet [75, 76]. Interventions to lose body fat mass have also been associated with changes in DNA methylation. Studies have reported that the DNA methylation profiles of adipose tissue [43, 64], peripheral blood mononuclear cells [77], and muscle tissue [78] in formerly obese patients become more similar to the profiles of lean subjects following weight loss. Weight loss surgery also partially reversed non-alcoholic fatty liver disease-associated methylation changes in liver [79] and in another study led to hypomethylation of multiple obesity candidate genes, with more pronounced effects in subcutaneous compared to omental (visceral) fat [64]. Accumulating evidence suggests that exercise interventions can also influence DNA methylation. Most of these studies have been conducted in lean individuals [80�82], but one exercise study in obese T2DM subjects also demonstrated changes in DNA methylation, including in genes involved in fatty acid and glucose transport [83]. Epigenetic changes also occur with aging, and recent data suggest a role of obesity in augmenting them [9, 84, 85]. Obesity accelerated the epigenetic age of liver tissue, but in contrast to the findings described above, this effect was not reversible after weight loss [84].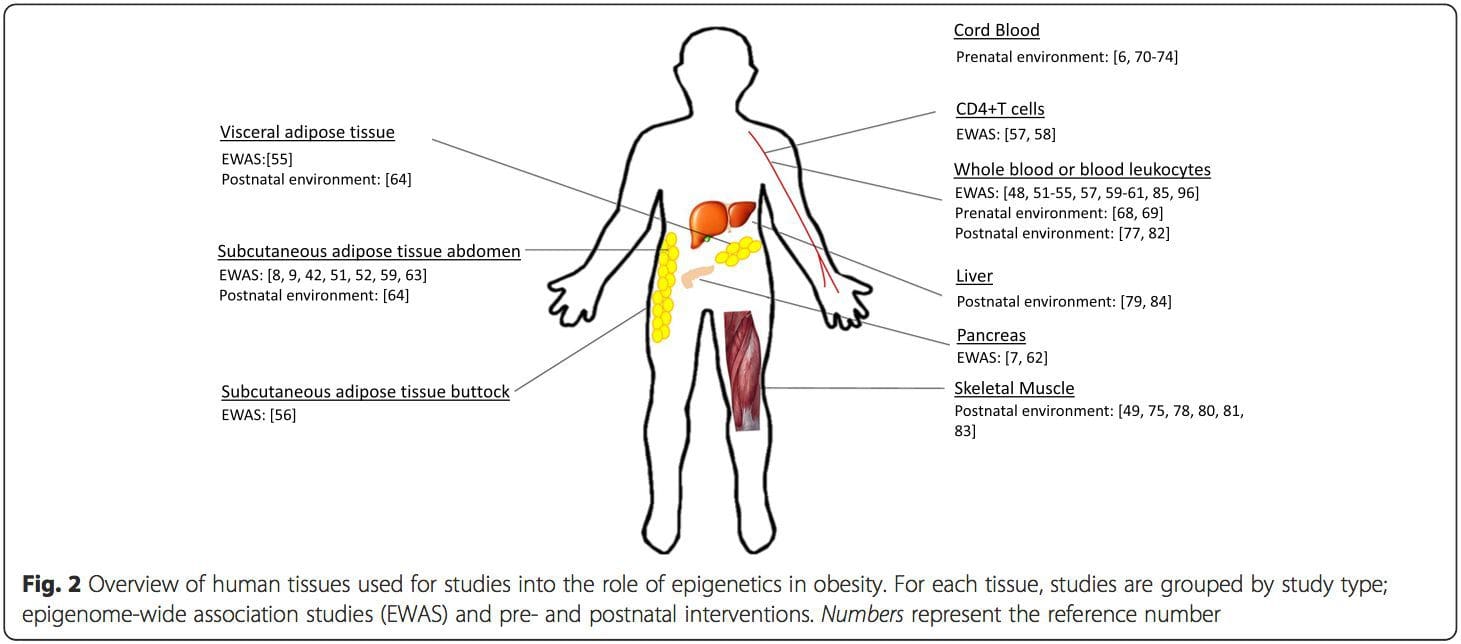 faaiuga
faaiuga



